|
|
本帖最后由 金鹏 于 2016-8-24 23:02 编辑
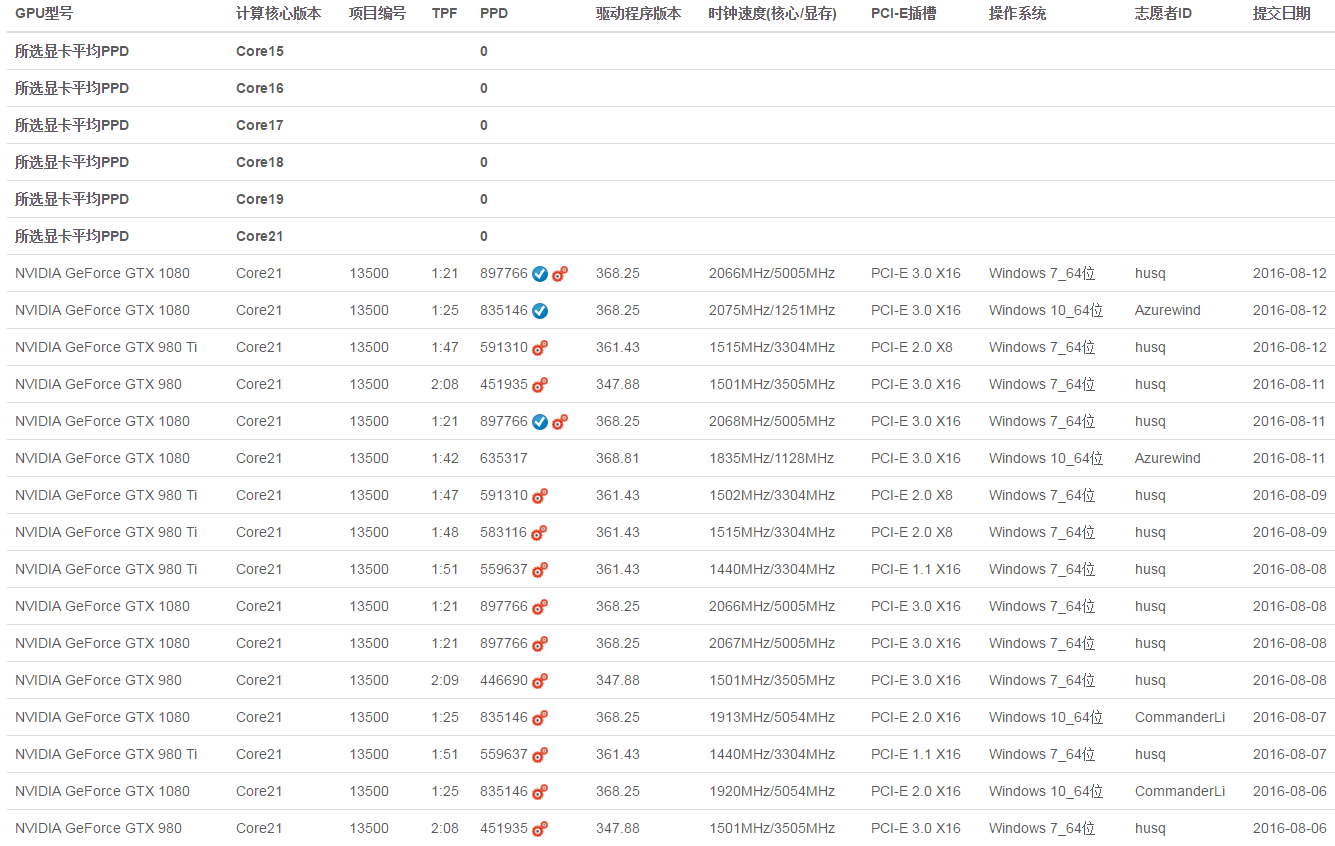

P13500 is moved to FAH[size=1.3em]Project 13500 is now in FAH.
number of atoms: 36948
credit: 9410
k-factor: 0.75
timeout: 7
deadline: 10
13500: One of the challenges of predicting the binding affinity of small molecules to protein targets is conformational flexibility of the protein. In this project, we will simulate T4 Lysozyme L99A protein as a model to study the effect of protein binding site flexibility and contribution of reorganization energy to binding energy. T4 Lysozyme L99A has been a key model system in understanding small drug binding to proteins and testing methods of free energy of binding predictions. Starting structures of T4 Lysozyme L99A in this project come from ligand bound crystal structures of this protein in a growing ligand series that highlights adaptation of protein conformation depending on ligand binding.
Thanks for folding! P13500 moving to ADVANCED[size=1.3em]New project 13500 is now in ADVANCED.
number of atoms: 36948
credit: 9410
k-factor: 0.75
timeout: 7
deadline: 10
13500: One of the challenges of predicting the binding affinity of small molecules to protein targets is conformational flexibility of the protein. In this project, we will simulate T4 Lysozyme L99A protein as a model to study the effect of protein binding site flexibility and contribution of reorganization energy to binding energy. T4 Lysozyme L99A has been a key model system in understanding small drug binding to proteins and testing methods of free energy of binding predictions. Starting structures of T4 Lysozyme L99A in this project come from ligand bound crystal structures of this protein in a growing ligand series that highlights adaptation of protein conformation depending on ligand binding.
Thanks for testing! Project 13500 [Core21, LIN/WIN, GPU][size=1.3em]Posting here for Mehtap Isik from Chodera Lab, while she waits for posting rights in the Beta forums!
Project 13500
By MehtapIsik
New project 13500 is now in BETA.
number of atoms: 36948
credit: 9410
k-factor: 0.75
timeout: 7
deadline: 10
13500: One of the challenges of predicting the binding affinity of small molecules to protein targets is conformational flexibility of the protein. In this project, we will simulate T4 Lysozyme L99A protein as a model to study the effect of protein binding site flexibility and contribution of reorganization energy to binding energy. T4 Lysozyme L99A has been a key model system in understanding small drug binding to proteins and testing methods of free energy of binding predictions. Starting structures of T4 Lysozyme L99A in this project come from ligand bound crystal structures of this protein in a growing ligand series that highlights adaptation of protein conformation depending on ligand binding.
Thanks for testing!
|
|