 |
|
|
 |


FightAIDS@Home |
 |

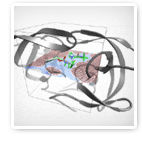 UNAIDS, the Joint United Nations Program on HIV/AIDS, estimated that in 2004 there were more than 40 million people around the world living with HIV, the Human Immunodeficiency Virus. The virus has affected the lives of men, women and children all over the world. Currently, there is no cure in sight, only treatment with a variety of drugs.
UNAIDS, the Joint United Nations Program on HIV/AIDS, estimated that in 2004 there were more than 40 million people around the world living with HIV, the Human Immunodeficiency Virus. The virus has affected the lives of men, women and children all over the world. Currently, there is no cure in sight, only treatment with a variety of drugs.
Prof. Arthur J. Olson's laboratory at The Scripps Research Institute (TSRI) is studying computational ways to design new anti-HIV drugs based on molecular structure. It has been demonstrated repeatedly that the function of a molecule — a substance made up of many atoms — is related to its three-dimensional shape. Olson's target is HIV protease ("pro-tee-ace"), a key molecular machine of the virus that when blocked stops the virus from maturing. These blockers, known as "protease inhibitors", are thus a way of avoiding the onset of AIDS and prolonging life. The Olson Laboratory is using computational methods to identify new candidate drugs that have the right shape and chemical characteristics to block HIV protease. This general approach is called "Structure-Based Drug Design", and according to the National Institutes of Health's National Institute of General Medical Sciences, it has already had a dramatic effect on the lives of people living with AIDS.
Even more challenging, HIV is a "sloppy copier," so it is constantly evolving new variants, some of which are resistant to current drugs. It is therefore vital that scientists continue their search for new and better drugs to combat this moving target.
Scientists are able to determine by experiment the shapes of a protein and of a drug separately, but not always for the two together. If scientists knew how a drug molecule fit inside the active site of its target protein, chemists could see how they could design even better drugs that would be more potent than existing drugs.
To address these challenges, World Community Grid's FightAIDS@Home project runs a software program called AutoDock developed in Prof. Olson's laboratory. AutoDock is a suite of tools that predicts how small molecules, such as drug candidates, might bind or "dock" to a receptor of known 3D structure. The very first version of AutoDock was written in the Olson Laboratory in 1990 by Dr. David S. Goodsell, while newer versions, developed by Dr. Garrett M. Morris, have been released which add new scientific understanding and strategies to AutoDock, making it computationally more robust, faster, and easier for other scientists to use. AutoDock is used on the World Community Grid to dock large numbers of different small molecules to HIV protease, so the best molecules can be found computationally, selected and tested in the laboratory for efficacy against the virus, HIV. By joining forces together, The Scripps Research Institute, World Community Grid and its growing volunteer force can find better treatments much faster than ever before.
|

 |
 |
|
|
 |
